Publications/Preprints
2025
2024
2021
6. Amanda Lanza, John Blazeck, Nathan Crook, and Hal Alper, 2012. Linking yeast Gcn5p catalytic function and gene regulation using a quantitative, graded dominant mutant approach. PLoS ONE 7(4), e36193. Link to paper
5. Amanda Lanza, Nathan Crook, and Hal Alper, 2012. Innovation at the Intersection of Synthetic and Systems Biology. Current Opinion in Biotechnology 23(5), 712-717. Link to paper
16. Aura Ferreiro*, Nathan Crook*, Andrew J. Gasparrini, and Gautam Dantas, 2018. Multiscale Evolutionary Dynamics of Host-Associated Microbiomes. Cell, 172(6), 1216-1227. Link to paper
2016
15. Nathan Crook*, Jie Sun*, Joe Abatemarco*, James Wagner, Alexander Schmitz, and Hal Alper, 2016. in vivo continuous evolution of genes and pathways in yeast. Nature Communications, 7:13051. Link to paper
14. Amy Langdon*, Nathan Crook*, Gautam Dantas, 2016. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Medicine 8(39). Link to paper
13. Nathan Crook, Jie Sun, Nicholas Morse, Alexander Schmitz, and Hal Alper, 2016. Identification of gene knockdown targets conferring enhanced isobutanol and 1-butanol tolerance to Saccharomyces cerevisiae using a tunable RNAi screening approach. Applied Microbiology and Biotechnology, 100(23), 10005-10018. Link to paper
12. Jinsuk Lee*, Nathan Crook*, Jie Sun, and Hal Alper, 2016. Improvement of lactic acid production by a deletion of ssb1 in Saccharomyces cerevisiae. Journal of Industrial Microbiology & Biotechnology 43(1), 87-96. Link to paper
11. Kate Curran*, Nathan Crook*, Ashty Karim, Akash Gupta, and Hal Alper, 2014. Model-based design of synthetic yeast promoters via tuning of nucleosome architecture. Nature Communications, 5:4002. Link to paper
10. Nathan Crook, Alexander Schmitz, and Hal Alper, 2014. Optimization of a yeast RNA interference system for controlling gene expression and enabling rapid metabolic engineering. ACS Synthetic Biology 3(5), 307–313. Link to paper
9. Nathan Crook and Hal Alper, 2013. Model-Based Design of Synthetic, Biological Systems. Chemical Engineering Science 103, 15 November 2013, 2-11. Link to paper
8. Nathan Crook and Hal Alper, 2013. Classical Strain Improvement. Engineering Complex Phenotypes in Industrial Strains, First Edition 1-33. Edited by Ranjan Patnaik. John Wiley & Sons. Link to paper
7. Eric M. Brustad, Victor S. Lelyveld, Christopher D. Snow, Nathan C. Crook, Sang Taek Jung, Francisco M. Martinez, Timothy J. Scholl, Alan Jasanoff, Frances H. Arnold, 2012. Structure-Guided Directed Evolution of Highly Selective P450-Based Magnetic Resonance Imaging Sensors for Dopamine and Serotonin. Journal of Molecular Biology 422(2), 245-262. Link to paper
2018
4. Kate Curran, Nathan Crook, and Hal Alper, 2012. Using Flux Balance Analysis to Guide Microbial Metabolic Engineering. Methods in Molecular Biology v. 834: Microbial Metabolic Engineering 197-216. Link to paper
3. Nathan Crook, Elizabeth Freeman, and Hal Alper, 2011. Re-Engineering Multicloning Sites for Function and Convenience. Nucleic Acids Research 39(14), e92. Link to paper
2. Rudi Fasan, Nathan C. Crook, Matthew W. Peters, Peter Meinhold, Thomas Buelter, Marco Landwehr, Patrick C. Cirino, and Frances H. Arnold, 2011. Improved Product-Per-Glucose Yields in P450-Dependent Propane Biotransformations Using EngineeredEscherichia coli. Biotechnology and Bioengineering 108(3), 500-510. Link to paper

2014

2013

2012




2011

2019
17. Nathan Crook*, Aura Ferreiro*, Andrew J. Gasparrini, Mitchell W. Pesesky, Molly K. Gibson, Bin Wang, Xiaoqing Sun, Zevin Condiotte, Stephen Dobrowolski, Daniel Peterson, and Gautam Dantas, 2019. Adaptive Strategies of the Candidate Probiotic E. coli Nissle in the Mammalian Gut.
Cell Host & Microbe https://doi.org/10.1016/j.chom.2019.02.005
Highlights:
-
Can the Microbiome Deliver? A Proof-of-Concept Engineered E. coli PKU Therapeutic CellPress
18. Justin M. Vento, Nathan Crook, Chase L. Beisel, 2019. Barriers to genome editing with CRISPR in bacteria. J Ind Microbiol Biotechnol.
2020
19. Nathan Crook, Aura Ferreiro, Zevin Condiotte, Gautam Dantas, 2020. Transcript Barcoding Illuminates the Expression Level of Synthetic Constructs in E. coli Nissle Residing in the Mammalian Gut. ACS SynBio
20. Deniz Durmusoglu*, Ibrahim Al’Abri*, Scott P. Collins, Junrui Cheng, Abdulkerim Eroglu, Chase Beisel, Nathan Crook, 2020. In Situ Biomanufacturing of Small Molecules in the Mammalian Gut by Probiotic Saccharomyces boulardii. ACS SynBio
21. Ibrahim Al’Abri, Deniz Durmusoglu, Nathan Crook, 2021. What E. coli knows about your 1-year-old infant: Antibiotic use, lifestyle, birth mode, and siblings. Cell Host & Microbe

22. Deniz Durmusoglu*, Carly M Catella*, Ethan F Purnell*, Stefano Menegatti, Nathan Crook, 2021. Design and in situ biosynthesis of precision therapies against gastrointestinal pathogens. Current Opinion in Physiology

23. Mairead K. Heavey*, Deniz Durmusoglu*, Nathan Crook, Aaron C. Anselmo, 2021. Discovery and delivery strategies for engineered live biotherapeutic products. Trends in Biotechnology

24. Ki Yoon Kwon, Samuel Cheeseman, Alba Frias-DeDiego, Haeleen Hong, Jiayi Yang, Woojin Jung,Hong Yin, Billy J. Murdoch, Frank Scholle, Nathan Crook, Elisa Crisci, Michael D. Dickey, Vi Khanh Truong, Tae-il Kim, 2021.
A Liquid Metal Mediated Metallic Coating for Antimicrobial and Antiviral Fabrics. Advanced Materials

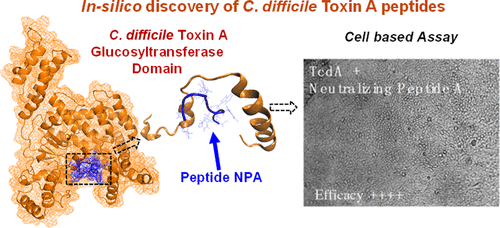
25. Xingqing Xiao, Sudeep Sarma, Stefano Menegatti, Nathan Crook, Scott T. Magness, and Carol K. Hall 2021.
In Silico Identification and Experimental Validation of Peptide-Based Inhibitors Targeting Clostridium difficile Toxin A. ACS Chem. Biol
26. Emil D. Jensen, Marcus Deichmann, Xin Ma, Rikke U. Vilandt, Giovanni Schiesaro, Marie B. Rojek, Bettina Lengger, Line Eliasson, Justin M. Vento, Deniz Durmusoglu, Sandie P. Hovmand, Ibrahim Al’Abri, Jie Zhang, Nathan Crook, Michael K. Jensen 2022.
Engineered cell differentiation and sexual reproduction in probiotic and mating yeasts. Nature Communications
2022
27. Ibrahim S Al'Abri, Daniel Haller, Zidan Li, Nathan C Crook, 2022. Inducible Directed Evolution of Complex Phenotypes in Bacteria. Nucleic Acids Research
28. Suryang Kwak *, Nathan Crook *, Aki Yoneda, Naomi Ahn, Jie Ning, Jiye Cheng, Gautam
Dantas, 2022. Functional mining of novel terpene synthases from metagenomes. Biotechnology for Biofuels and Bioproducts

29. Ibrahim S Al’Abri*, Zidan Li*, Daniel J Haller*, Nathan Crook, 2022. A Novel Method of Inducible Directed Evolution to Evolve Complex Phenotypes. Bio-protocol
30. Abdulkerim Eroglu, Ibrahim Al’Abri, Rachel E. Kopec, Nathan Crook, Torsten Bohn, 2023. Carotenoids and their health benefits as derived via their interactions with gut microbiota. Advances in Nutrition 14(2) 238-255


31. Rachel S. Bang, Michael Bergman, Tianyu Li, Fiona Mukherjee, Abdulelah S. Alshehri, Nicholas L. Abbott, Nathan C. Crook, Orlin D. Velev, Carol K. Hall, Fengqi You, 2023. An integrated chemical engineering approach to understanding microplastics. AiChE Journal 69(4) e18020

32. Deniz Durmusoglu, Ibrahim Al’Abri, Zidan Li, Taufika Islam Williams, Leonard B. Collins, José L. Martínez, Nathan Crook, 2023. Improving therapeutic protein secretion in the probiotic yeast Saccharomyces boulardii using a multifactorial engineering approach. Microbial Cell Factories

34.Tianyu Li, Stefano Menegatti, Nathan Crook Breakdown of polyethylene therepthalate microplastics under saltwater conditions using engineered Vibrio natriegens. AIChE Journal. 2023; e18228

35.Torsten Bohn, Emilio Balbuena, Hande Ulus, Mohammed Iddir, Genan Wang, Nathan Crook, Abdulkerim Eroglu. Carotenoids in health as studied by omics-related endpoints. Advances in Nutrition.

36. John van Schaik, Zidan Li, John Cheadle, Nathan Crook,
2023. Engineering the Maize Root Microbiome: A Rapid MoClo Toolkit and Identification of Potential Bacterial Chassis for Studying Plant–Microbe Interactions. ACS Synthetic Biology

1. Rudi Fasan, Michael M. Chen, Nathan C. Crook and Frances H. Arnold, 2007. Engineered Alkane-Hydroxylating Cytochrome P450BM3 Exhibiting Native-like Catalytic Properties. Angewandte Chemie International Edition 46(44), 8414-8418. Link to paper
2007

37. Mariam Sohail, Tahira Pirzada, Richard Guenther, Eduardo Barbieri, Tim Sit, Stefano Menegatti, Nathan Crook, Charles H. Opperman, and Saad A. Khan. 2023. Cellulose Acetate-Stabilized Pickering Emulsions: Preparation, Rheology, and Incorporation of Agricultural Active Ingredients. ACS Sustainable Chem. Eng.

NeNeN
40. Justin M. Vento, Deniz Durmusoglu, Tianyu Li, Constantinos Patinios, Sean Sullivan, Fani Ttofali, John van Schaik, Yanying Yu, Yanyan Wang, Lars Barquist, Nathan Crook, and Chase L. Beisel, 2024. A cell-free transcription-translation pipeline for recreating methylation patterns boosts DNA transformation in bacteria. Molecular Cell

38. Zidan Li, Nathan Crook. 2023. Chips, guts, and gas: unraveling volatile microbial mysteries in real time! Trends in Biotechnology

2023
39. Deniz Durmusoglu, Daniel J. Haller, Ibrahim S. Al’Abri, Katie Day, Carmen Sands, Andrew Clark, Adriana San-Miguel, Ruben Vazquez-Uribe, Morten O. A. Sommer, and Nathan Crook, 2024. Programming Probiotics: Diet-Responsive Gene Expression and Colonization Control in Engineered S. boulardii. ACS Synthetic Biology

41. Ethan Gates, Nathan Crook, 2024. The biochemical mechanisms of plastic biodegradation. FEMS Microbiology Reviews
.png)
42. Mariam Sohail, John Cheadle, Rishum Khan, Hrishikesh Mane, Khandoker Samaher Salem, Katie Ernst, Adriana San Miguel, Charles H. Opperman, Tahira Pirzada, Nathan Crook, Saad A. Khan, 2025. Pickering Emulsion for Enhanced Viability of Plant Growth Promoting Bacteria and Combined Delivery of Agrochemicals and Biologics. Advanced Functional Materials

43. William Parker, Amanda Taylor, Aryan Razdan, Jose Escarce, Nathan Crook, 2025. Enabling technologies for in situ biomanufacturing using probiotic yeast. Advanced Drug Delivery Reviews

41. Abdulkerim Eroglu, Genan Wang, Nathan Crook, Torsten Bohn, 2024. Carotenoids. Advances in Nutrition

